Exploring antibody validation approaches with AbVal meeting speakers: Cell Signaling Technology
5
Min Read
In this blog:
- We speak to AbVal meeting speaker Antony Wood, Senior Director, Product Design & Strategy, Cell Signaling Technology, about their validation approach
- The journey of an antibody from development to use is followed
- We delve into how CST place importance on validating their antibodies in specific applications
As we approach the 4th International Antibody Validation meeting, we will be talking to our sponsors who are speaking at the meeting to understand a little more about their validation strategies ahead of their talks this September.
Today, we focus on the validation approach of Cell Signaling Technology.
We are guided through the process of validating their antibodies for specificity in each application independently. CST uses six hallmarks of antibody validation in this process, leading to antibodies being passed or failed for specific assays.
The validation journey of a Cell Signaling Technology antibody
To better understand Cell Signaling Technology’s antibody validation approach, we follow an example: two 53BP1 clones. We were taken through this journey by Katie Crosby, Senior Director, Antibody Applications & Validation, at CST.
Antibodies are developed for the desired applications
Ahead of development, teams at CST will look into how a target is being studied or could be studied. Application testing is then determined based on this information, to ensure validation is being carried out for the most relevant applications. In the examples we discuss today for the 53BP1 clones, IHC and WB were chosen.
Hallmarks of antibody validation
Cell Signaling Technology have coined the term the ‘hallmarks of antibody validation’ to describe the six complementary strategies they use to test for antibody specificity in a given application. These are:
- Binary Strategy
- Ranged Strategy
- Orthogonal Strategy
- Multiple Antibody Strategy
- Heterologous Strategy
- Complementary Strategy
Different teams of application specialists will carry out the strategies, with the method used often depending on the application, biological context of the target, model availability or other factors.
Validating antibodies for specificity in each application
For the two 53BP1 clones, a knockout sample tested for western blotting gave evidence that each antibody was specific, meaning the clones passed for this application. However, when tested for IHC it was found that one of the clones, E9I7E, displayed a non-specific signal at the cell membrane. This highlights a significant challenge in antibody validation – KO validation in WB cannot predict IHC performance.
–
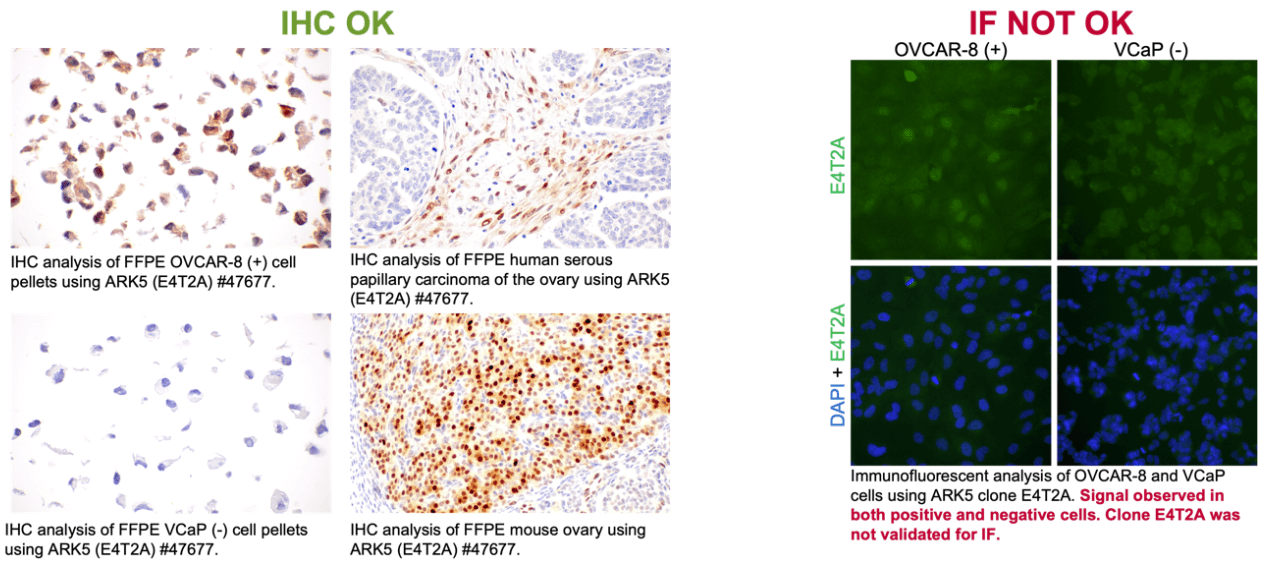
CST provided further results from their validation which showed how specificity in IHC does not imply the same in IF-IC, and vice versa. Specifically, they shared an experiment where the ARK5 (E4T2A) antibody was validated for IHC, but failed IF testing. In paraffin-embedded cell pellets, E4T2A demonstrated a signal in a positive cell line but not in a negative cell line, whereas IF-IC testing in those same models failed to yield the same binary results. Further testing in paraffin-embedded human tissues with this clone yielded the appropriate signal with no appreciable background, consistent with the requirements for a recommendation for use in IHC-P.
–
Similarly, the Claudin-6 (E2S5M) Rabbit mAb antibody was validated for IF-IC after demonstration of an appropriate signal at the cell membrane in a positive cell line with no signal observed in a negative cell line. When those same paraffin-embedded cell lines were tested by IHC-P, limited specific signal in the positive cell line was observed along with prominent non-specific nuclear signal in both cell lines. Titration of the antibody failed to eliminate the non-specific signal in the positive cell line, leaving minimal specific signal. Despite appropriate functionality in an IF-IC assay, this clone was not recommended for IHC-P.
–
Candidates pass or fail based on data
After applying these strategies, validation data are evaluated in an application-specific manner, and clones are passed or failed for each application independently. Or, in other words, ‘performance in one application neither guarantees nor requires performance in another’.
Below, we provide a summary of the antibody validation examples discussed:
–
Data are made available to researchers so they can also evaluate why antibodies have been passed or failed for certain applications – giving them more confidence that the antibody will work specifically for their chosen experiment.
Interested in learning more about antibody validation?
We can’t wait for the 4th International Antibody Validation meeting to learn even more about current validation practices, the future of the field, its challenges and more. With experts from academia, the pharma/biotech sector and antibody suppliers, it’s lining up to be a really exciting event.
Visit the website now to learn more and register: https://www.antibodyvalidation.co.uk/
Registration closes: 31st August 2023
At the Antibody Validation meeting, Antony Wood will be speaking for CST with a talk titled: ‘De-risking the data – The critical importance of validation in the age of multiplex and spatial biology’.
If you have interesting validation to share with us, please do reach out! We are always looking to learn more about antibody validation and keen to share that with our users.
- Skye and the CiteAb team